Overview
A natural product database for compound screening and other applications in natural compound research
Originally developed by Dr. Hartmut Laatsch, the renowned AntiBase Library is a comprehensive natural products library of more than 105,300 compounds. And with the latest release we've added over 9,500 more compounds!
Widely cited in scientific literature, this growing database is an effective screening tool for metabolomics and drug discovery where natural products and derivatives can accelerate treatments in for cancer, diabetes, disease, and antimicrobial or antifungal research.
Use the included Wiley's ChemWindow software to securely search the collection, plus powerful tools build your own databases, draw structures, and more.
Comprehensive coverage. |
Carefully curated data. |
Powerful data mining tools. |
|
Faster discovery with expanded compound coverage across various categories in this rich resource for natural product research. |
At Wiley, we prioritize data quality for results you can rely on to make informed decisions. |
Robust data mining tools to gain deeper insights, including the ability to build and incorporate your own data, and more! |
Applications
- Incorporate in high-throughput screening (HTS) and dereplication workflows in pharma and biotech to quickly identify compounds already studied to streamline the search for novel compounds having antimicrobial or desired effects.
- Use throughout various industries such as food & cosmetics, agriculture, pesticides, and other applications where natural products are prevalent or in academic programs and research.
What's New in the 2025 Release
We've increased the collection by ~10% with the addition of over 9,500 new compounds, including compounds from the latest release of Natural Product Atlas.
Trusted Data from a Trusted Source
Wiley is an authoritative source for scientific data. Our renowned databases are processed according to rigorous protocols to ensure they are of the highest quality. Qualification procedures start at data acquisition and continue throughout the database development process. Any data acquired from trusted partners is thoroughly vetted before inclusion in our collections.
Key Features
| Carefully curated and verified data | Data is collected from primary and secondary literature; then carefully checked and validated with key references and sources provided as a data field. |
| Valuable metadata to help you narrow results |
In addition to chemical structures, records contain searchable metadata where available such as:
|
| Spectral data to aid in characterization |
|
| Powerful tools for data mining and databasing |
Includes Wiley’s ChemWindow software to search the collection, plus powerful tools build your own databases, draw structures, and more. |
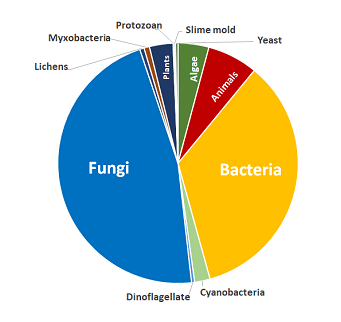
Compound Coverage
Includes compounds from these biological sources:
- Algae
- Animals
- Bacteria
- Cyanobacteria
- Dinoflagellate
- Fungi
- Lichens
- Myxobacteria
- Plants
- Protozoan
- Slime mold
- Yeast
Library Specifications
|
|
|
Use Cases from Literature
The AntiBase Library has been referenced over 1,500 times in Google Scholar. Here are a few of the many examples of how scientists use this database in various applications.
| Article Title | DOI Link | Summary | Application Area |
| Discovery of actinomycin L, a new member of the actinomycin family of antibiotics | https://doi.org/10.1038/s41598-022-06736-0 | Streptomycetes produce bioactive natural products, including a majority of naturally produced antibiotics. Two new actinomycin analogues produced by Streptomyces sp. MBT27, actinomycin L1 and L2, have been discovered. Dereplication was performed using Reaxys, ChemSpider, and AntiBase. Mass features of existing natural products were annotated while one mass feature with an m/z value of 1387.6706 [M + H]+ could not be matched to any previously reported natural products. | Antibiotics |
| New Metabolites From the Endophytic Fungus Cercophora samala Associated With Mitragyna inermis | https://doi.org/10.1177/1934578×211013220 | Mitragyna inermis is a small tree species that grows in West Africa that has been used in traditional medicine to treat a number of conditions. Two new metabolites from this tree species have been isolated and have had their structures elucidated by comparison of their spectroscopic data with those reported in AntiBase and by DFT calculations. | Metabolomics |
| Combination of high-throughput microfluidics and FACS technologies to leverage the numbers game in natural product discovery | https://doi.org/10.1111/1751-7915.13872 | To facilitate the discovery of natural products, a screening method was developed using a combination of droplet microfluidics and fluorescence-activated cell sorting-based technologies. This method was utilized to assess a microbial environmental sample. To identify the natural products, molecular formula searches were performed using AntiBase. | Antimicrobials |
| Screening for biologically active metabolites with endosymbiotic bacilli isolated from arthropods | https://doi.org/10.1111/j.1574-6968.2002.tb11475.x | Endosymbiotic bacteria from the genus Bacillus were isolated from different compartments of the gut of various insects and millipedes. The samples were screened for biologically active metabolites, which were isolated. Structure elucidation was done by a combination of NMR and HREI or DCI mass spectrometry. Their structures were determined with the aid of AntiBase, NMR: Aldrich Library, and spectrum collections (MS: NIST, WILEY). | Metabolomics |
| Comprehensive Analysis of the Alternaria Mycobolome Using Mass Spectrometry Based Metabolomics | https://doi.org/10.1002/mnfr.201900558 | Fourier-transform ion cyclotron resonance mass spectrometry (FTICR-MS) and LC-MS/MS are combined for the non-targeted and targeted analysis of the metabolome of three A. alternata isolates and one A. solani isolate. Due to the ultra-high resolution of FTICR-MS, unique molecular formulae are assigned to measured m/z signals. The molecular formulae are matched to entries of the databases AntiBase and Kyoto Encyclopedia of Genes and Genomes. The non-targeted analysis of the fungal extracts reveals variations in the secondary metabolite profile of A. alternata and A. solani. Differences in the biosynthesis of dibenzo-α-pyrones, perylene quinones, tentoxin, and tenuazonic acid of the A. alternata and A. solani isolates are determined applying targeted LC-MS/MS. | Metabolomics |
| Signalling and Bioactive Metabolites from Streptomyces sp. RK44 | https://doi.org/10.3390%2Fmolecules25030460 | Chemical profiling of the RK44 extract using high-resolution electrospray ionization mass spectrometry (HR-ESIMS) and Global Natural Product Social (GNPS) molecular network (MN) [7] showed a cluster of low molecular weight metabolites with a characteristic 250 nm UV absorption, suggesting that they share a common chromophore skeleton. Database dereplication using AntiBase 2017 (WILEY) and other online databases (The Natural Products Atlas, ChemSpider) [8,9] revealed that these metabolites have not been previously described in the literature [10,11]. | Metabolomics |
| A selective antibiotic for Lyme disease | https://doi.org/10.1016/j.cell.2021.09.011 | An extract of S. hygroscopicus was separated into 40 fractions using high-performance liquid chromatography (HPLC). The fractions were tested for growth inhibition of B. burgdorferi, resulting in the isolation of a pure compound with a molecular mass of 511 Da (Figure 1A). The structure was determined by a combination of mass spectrometry (MS) and NMR analysis (Figures S1 and S2; Tables S1 and S2). Surprisingly, a search of AntiBase database revealed the identity of this compound to hygromycin A, a known antibiotic (Figure 1A). | Antibiotics |
| Crude Extracts of Talaromyces Strains (Ascomycota) Affect Honey Bee (Apis mellifera) Resistance to Chronic Bee Paralysis Virus | https://doi.org/10.3390/v15020343 | Viruses are a large contributor to the global decline of honeybee populations. Introducing natural antiviral compounds from fungi to the diet of honeybees could limit the impact of such viruses. Seven strains of the fungal genus Talaromyces infected by chronic bee paralysis virus (CBPV) were examined. AntiBase 2017 and an in-house reference database were used to narrow down the molecular structure and to highlight compounds of interest. Three extracts (from strains B13, B18 and B30) were found to mitigate CBPV infections and increase the survival rate of bees. | Metabolomics |
| Sargassum horneri (Turner) C. Agardh Extract Regulates Neuroinflammation In Vitro and In Vivo | https://doi.org/10.3390/cimb44110367 | Studies suggest that S. horneri can help prevent osteoporosis and control cholesterol, blood pressure and hyperlipidemia and cosmetics and functional foods containing S. horneri are known to have anti-aging, anti-allergic and whitening effects. Thus, the anti-neuroinflammatory effects of S. horneri extract on microglia were investigated. The active ingredient in S. horneri was confirmed using AntiBase, previously reported data, and LC-MS/MS analysis. S. horneri was found to elicit anti-neuroinflammatory effects by inhibiting phosphorylation of p38 MAPK and NF-κB and inhibited astrocytes and microglia activation in LPS-challenged mice brain. | Anti-inflammatory |
| Fungal Species and Multi-Mycotoxin Associated with Post-Harvest Sorghum (Sorghum bicolor (L.) Moench) Grain in Eastern Ethiopia | https://doi.org/10.3390/toxins14070473 | Sorghum is the main staple food crop in developing countries, but sorghum grain quantity and quality are affected by contaminating fungi both in the field and post-harvest stages. Samples of fungal species and multi-mycotoxins associated with sorghum grain in the post-harvest stage were examined. The allocation of the toxins to different species was done using AntiBase, previously reported data, and investigations of the metabolite profile of pure cultures of toxigenic fungi. The study revealed that the sorghum grains were significantly contaminated with co-occurrences of several mycotoxins and thus the importance of farmers being trained on the improved management of sorghum production. | Mycotoxins |
Ordering Information
|
Product Code |
Product |
|
978EALDB04188 |
Wiley Identifier of Natural Products (AntiBase Library + ChemWindow Software) (Annual Subscription) |
|
978EALDB05529 |
Wiley Identifier of Natural Products (AntiBase Library + ChemWindow Software) (Annual Subscription Renewal) |
|
978EALDB04195 |
Wiley Identifier of Natural Products (AntiBase Library) (Annual Subscription)* |
|
978EALDB05512 |
Wiley Identifier of Natural Products (AntiBase Library) (Annual Subscription Renewal)* |
*Requires KnowItAll software
Compatibility
- The Wiley Identifier of Natural Products includes and is compatible with the KnowItAll ChemWindow Edition software.
- It can also be used with other editions of the KnowItAll software you are using.